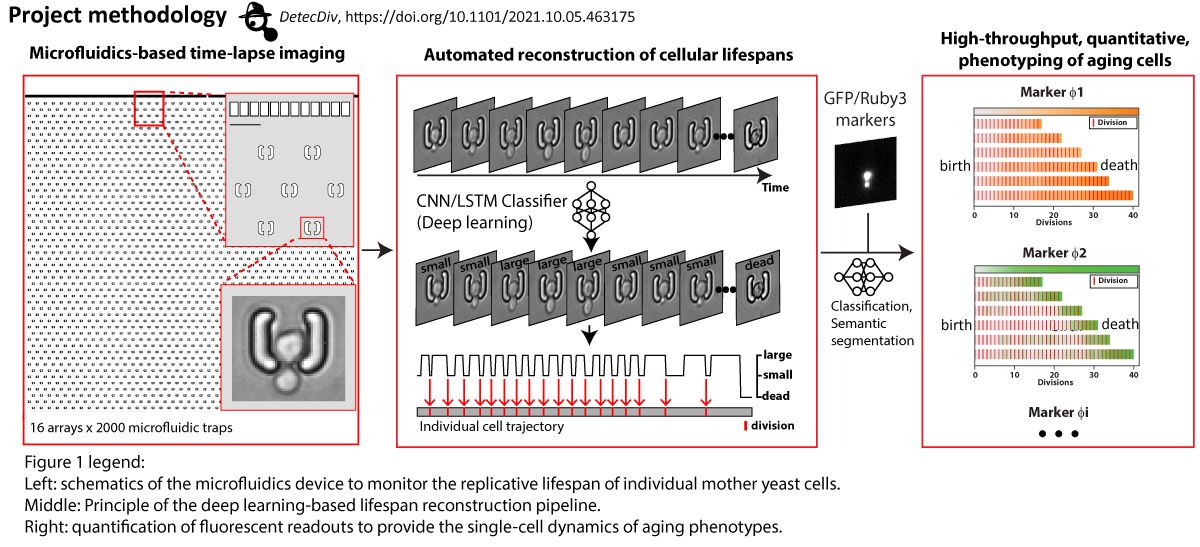
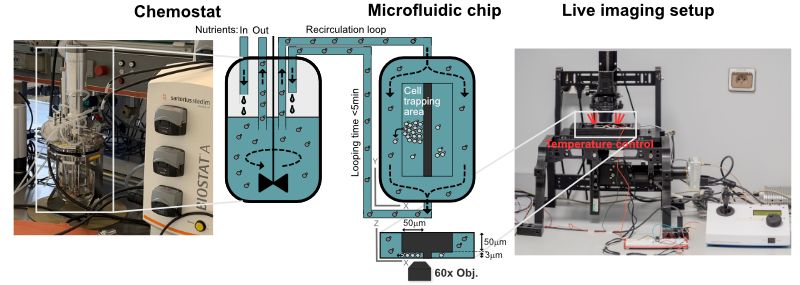
Under specific conditions of growth in a chemostat, yeast cell populations spontaneously synchronize to undergo sustained metabolic oscillations, which appear to be tightly coupled to cell growth and division. This entire oscillatory behavior is referred to as the yeast metabolic cycle (YMC). Intriguingly, our lab and others have recently shown that metabolic oscillations persist in the absence of cell division and even appear to be important in determining cell cycle entry and exit. Furthermore, we have shown that redox signaling is involved in the emergence of stable collective metabolic oscillatory behavior and in the coupling of the oscillatory metabolic state to cell division in the context of YMC. Recent studies reveal the existence of metabolic oscillations, redox signaling, and protein kinase A activity at the single cell level. Consistent with YMC, these oscillations may be coupled to cell division and appear to be important in regulating cell cycle entry and exit, but they may also persist independently of cell division. However, it is unclear whether the metabolic cycles observed at the single cell level correspond to those observed in CMJ-synchronized populations. The mechanistic basis of metabolic cycles in both systems is completely unknown and the exact causal relationships, interference and interaction between metabolic cycles and cell division remain elusive. In this thesis project, we will develop a new methodology that merges population-scale dynamic measurements with single-cell monitoring using microfluidics. By combining our expertise in yeast genetics, novel genetically encoded redox sensors, quantitative live cell imaging, microfluidics and chemostat cultures, we will elucidate population heterogeneity in terms of metabolic cycles and cell division with a spatial and temporal resolution previously unattainable. In addition, we will address the role of redox signaling and protein kinase A activity in coupling cell division with oscillatory metabolism. By combining our approaches with computational analyses, we will take important steps towards deciphering the fundamental principles that govern the emergence of metabolic oscillations, elucidating their potential fitness benefits in fluctuating environments, and understanding their relevance to cell division.