A Trade-Off between Stress Resistance and Tolerance Underlies the Adaptive Response to Hydrogen Peroxide
Basile Jacquel, Bor Kavčič, Audrey Matifas, Thomas Julou, Gilles Charvin Cell Systems 16, 101320 (2025) - DOI: 10.1016/j.cels.2025.101320
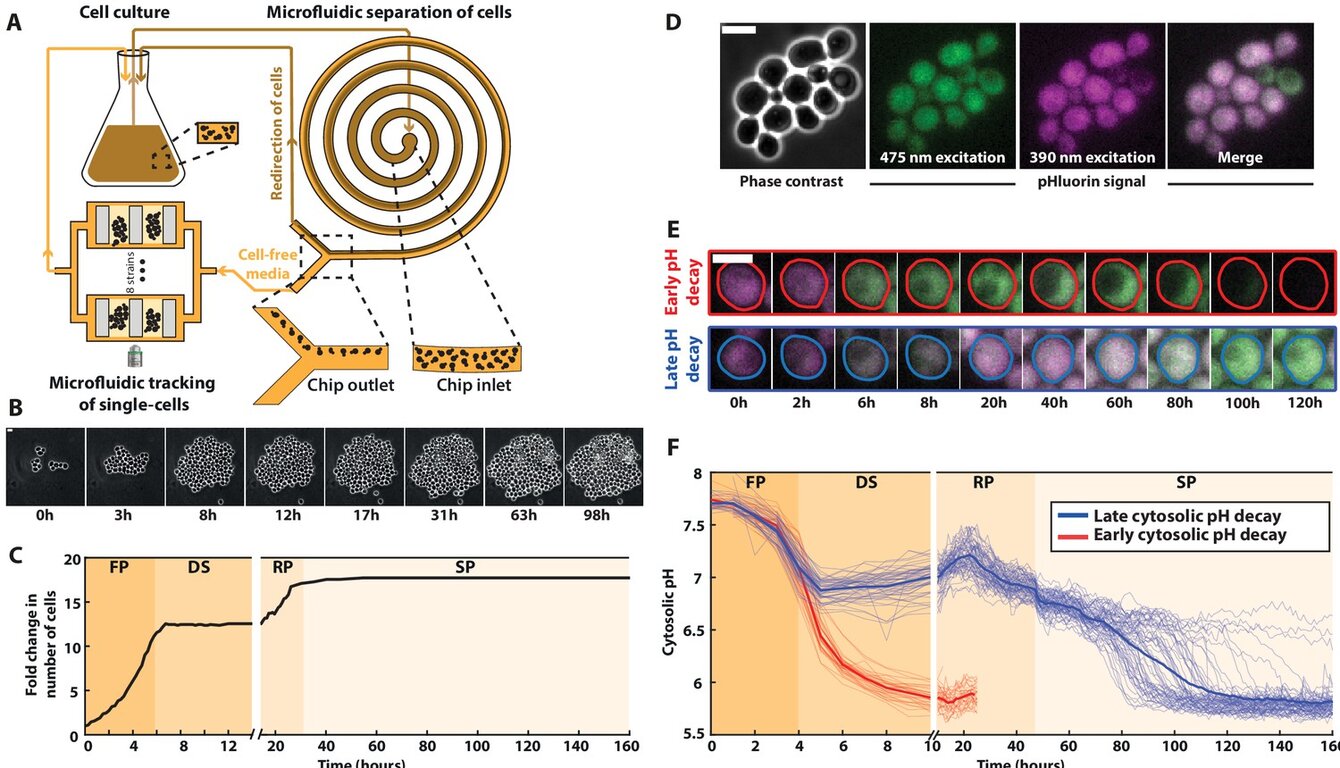
In this work in collaboration with the Thomas Julou and Bruce Morgan lab, we investigated the mechanistic interplay between resistance and tolerance strategies in Saccharomyces cerevisiae subjected to hydrogen peroxide (H₂O₂). Employing microfluidic cultivation and time-lapse fluorescence microscopy, we could quantitatively distinguish two discrete cellular responses: resistance, defined by the capacity to maintain proliferative growth via efficient detoxification of reactive oxygen species, and tolerance, characterized by a reversible proliferation arrest that prioritizes viability under excessive oxidative burden.
Through targeted deletion of ZWF1, the gene encoding the glucose-6-phosphate dehydrogenase of the pentose phosphate pathway responsible for NADPH production, we observed a pronounced decrease in H₂O₂ resistance concomitant with a paradoxical enhancement of tolerance. By combining additional genetic perturbations and varying extracellular nutrient conditions, we demonstrated that, upon overwhelming of the canonical redox homeostatic machinery, redox-dependent inhibition of protein kinase A (PKA) operates as a molecular switch. This inhibition redirects cells into a nutrient-dependent tolerant state, wherein proliferation is transiently suspended, thereby mitigating lethal oxidative damage.
Extending our analysis to Escherichia coli, our work revealed that the resistance–tolerance trade-off is conserved across phylogeny. These results established a unified framework in which redox signaling dynamically allocates cellular resources between detoxification and survival modes. By elucidating the regulatory logic governing this balance, our study provides a conceptual foundation for the development of combination therapies that concurrently target pathogen or tumor resistance mechanisms and tolerance defenses, with the objective of preventing treatment escape and disease relapse.
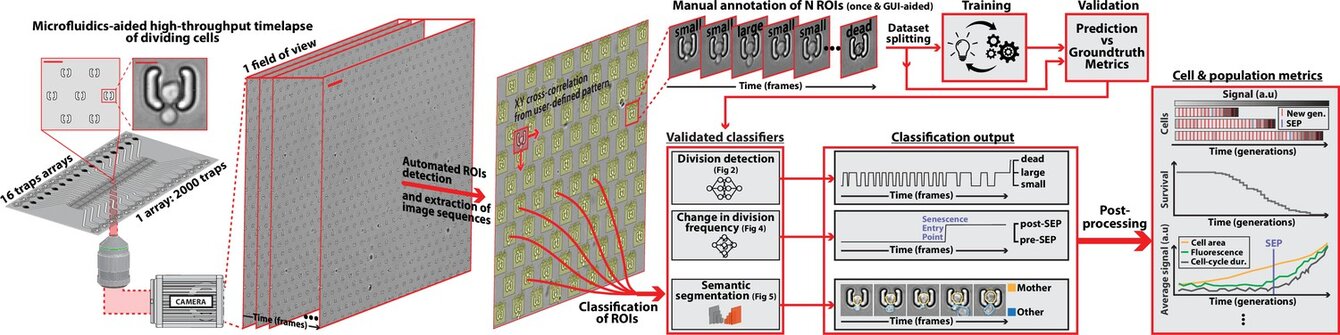
DetecDiv, a generalist deep-learning platform for automated cell division tracking and survival analysis
Théo Aspert, Didier Hentsch, Gilles Charvin. eLife, (2022) - DOI: 10.7554/eLife.79519
In this work, we used deep learning to classify sequences of images of individual yeast trapped in microfluidic cavities to auomatize their successive divisions.
We have shown that our analysis method allows for a very accurate measurement of the replicative life span of cells, or their survival in response to environmental stress. This method, which is very general, since it works very well on several trap geometries and imaging conditions, paves the way to more systematic studies of the mechanisms that control the entry into cellular senescence.
Monitoring single-cell dynamics of entry into quiescence during an unperturbed life cycle
Jacquel Basile, Aspert Théo, Laporte Damien, Sagot Isabelle, Charvin Gilles. eLife, 10:e73186, (2021) - DOI: 10.7554/eLife.73186
In this study, we developed a microfluidic system to monitor the entry into quiescence of individual yeast cells exposed to progressive nutrient depletion caused by population growth. We used fluorescent markers to characterize the dynamics of cell reorganization during different phases of quiescence entry. Our methodology reveals the emergence of a heterogeneity of cell fates, with some cells unable to enter respiration after the diauxic shift.
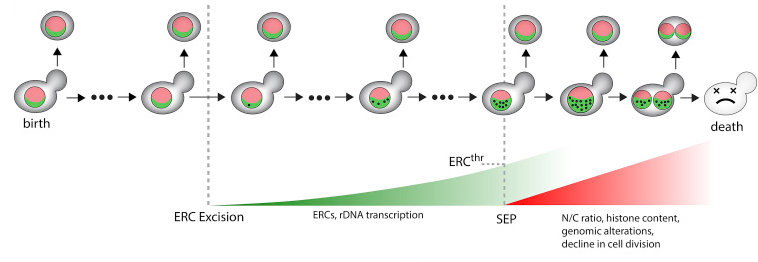
Excessive rDNA transcription drives the disruption in nuclear homeostasis during entry into senescence in budding yeast
Morlot S, Song J, Léger I, Matifas A, Gadal O, Charvin G. Cell Rep, (28)2:408-422, (2019) - DOI: 10.1016/j.celrep.2019.06.032
Dans ce travail, nous avons mesuré la dynamique de l'entrée en séensecence dans des cellules individuelles de levure. En particulier, nous avons démontré expérimentalement que les mini-cercles d'ADN ribosomiques extra-chromosomaux (ou ERCs), qui sont étroitement associées au vieillissement chez la levure, s'accumulent exponentiellement bien avant que que les cellules entrent en sénescence et conduisent à une accumulation massive d'ARN ribosomique dans le nucléole et de dysfonctionnements nucléaires majeurs. Nos expériences permettent ainsi de construire un scénario temporel quantitatif conduisant à l'entrée en sénescence.
2025
A Trade-Off between Stress Resistance and Tolerance Underlies the Adaptive Response to Hydrogen Peroxide. Basile Jacquel, Bor Kavčič, Audrey Matifas, Thomas Julou, Gilles Charvin Cell Systems 16, 101320 doi: 10.1016/j.cels.2025.101320
High-Throughput Measurement of Single-Fission Yeast Cell Volume Using Fluorescence Exclusion. Venkova L, García-Ruano D, Jain A, Charvin G, Coudreuse D. Methods Mol Biol. 2025;2862:7-32. doi: 10.1007/978-1-0716-4168-2_2. PMID: 39527190
2022
Left-right asymmetry in oxidative stress sensing neurons in C. elegans. Quintin S, Charvin G. MicroPubl Biol. (2022) - DOI: 10.17912/micropub.biology.000652. eCollection 2022.
Distinct mechanisms underlie H2O2 sensing in C. elegans head and tail Quintin Sophie, Aspert Théo, Charvin Gilles PLOS One, (2022) - DOI: 10.1371/journal.pone.0274226
DetecDiv, a deep-learning platform for automated cell division tracking and replicative lifespan analysis Théo Aspert, Didier Hentsch, Gilles Charvin eLife, (2022) - DOI: 10.7554/eLife.79519
A Microfluidic Platform for Tracking Individual Cell Dynamics during an Unperturbed Nutrients Exhaustion Théo Aspert, Basile Jacquel, Gilles Charvin Bio Protoc (2022) - DOI: 10.21769/BioProtoc.4470
Nuclear Pore Complex Acetylation Regulates mRNA Export and Cell Cycle Commitment in Budding Yeast Mercè Gomar-Alba, Vasilisa Pozharskaia, Celia Schaal, Arun Kumar, Basile Jacquel, Gilles Charvin, J. Carlos Igual, Manuel Mendoza EMBO J, (2022) - DOI: 10.15252/embj.2021110271
DNA circles promote yeast ageing in part through stimulating the reorganization of nuclear pore complexes Anne C. Meinema*, Anna Marzelliusardottir*, Mihailo Mirkovic*, Théo Aspert, Sung Sik Lee, Gilles Charvin, Yves Barral eLife, 11:e71196, (2022) - DOI: 10.7554/eLife.71196
2021
Monitoring single-cell dynamics of entry into quiescence during an unperturbed lifecycle Jacquel Basile, Aspert Théo, Laporte Damien, Sagot Isabelle, Charvin Gilles eLife, 10:e73186, (2021) - DOI: 10.7554/eLife.73186
Increased levels of mitochondrial import factor Mia40 prevent the aggregation of polyQ proteins in the cytosol Anna M Schlagowski, Katharina Knöringer, Sandrine Morlot, Ana Sánchez Vicente, Tamara Flohr, Lena Krämer, Felix Boos, Nabeel Khalid, Sheraz Ahmed, Jana Schramm, Lena M Murschall, Per Haberkant, Frank Stein, Jan Riemer, Benedikt Westermann, Ralf J Braun, Konstanze F Winklhofer, Gilles Charvin, Johannes M Herrmann EMBO J, (2021) - DOI: 10.15252/embj.2021107913
A trade-off between stress resistance and tolerance underlies the adaptive response to hydrogen peroxide Jacquel Basile, Matifas Audrey, Charvin Gilles bioRxiv, (2021) - DOI: 10.1101/2021.04.23.440814
2020
Single-Cell Tracing Dissects Regulation of Maintenance and Inheritance of Transcriptional Reinduction Memory Poonam Bheda, Diana Aguilar-Gómez, Nils B Becker, Johannes Becker, Emmanouil Stavrou, Igor Kukhtevich, Thomas Höfer, Sebastian Maerkl, Gilles Charvin, Carsten Marr, Antonis Kirmizis, Robert Schneider Mol. Cell, (78)5:915-925, (2020) - DOI: 10.1016/j.molcel.2020.04.016
2019
Proteostasis collapse halts G1 progression and delimits replicative lifespan Moreno D F, Jenkins K, Morlot S, Charvin G, Csikász-Nagy A, Aldea M eLife, 8:e48240, (2019) - DOI: 10.7554/eLife.48240
Excessive rDNA transcription drives the disruption in nuclear homeostasis during entry into senescence in budding yeast Morlot S, Song J, Léger I, Matifas A, Gadal O, Charvin G Cell Rep, (28)2:408-422, (2019) - DOI: 10.1016/j.celrep.2019.06.032
COSPLAY: An expandable toolbox for combinatorial and swift generation of expression plasmids in yeast Goulev Y, Matifas A, Heyer V, Reina-San-Martin B, Charvin G PLoS ONE, 14(8): e0220694, (2019) - DOI: 10.1371/journal.pone.0220694
Self-Learning Microfluidic Platform for Single-Cell Imaging and Classification in Flow Constantinou I, Jendrusch M, Aspert T, Görlitz F, Schulze A, Charvin G, Knop M. Micromachines, 10: 311, (2019) - DOI: 10.3390/mi10050311
2018
Adaptation to DNA damage checkpoint in senescent telomerase-negative cells promotes genome instability Coutelier H, Xu Z, Morisse MC, Lhuillier-Akakpo M, Pelet S, Charvin G, Dubrana K, Teixeira MT. Genes Dev., 1;32(23-24):1499-1513, (2018) - DOI: 10.1101/gad.318485.118
Multiple inputs ensure yeast cell size homeostasis during cell cycle progression Garmendia-Torres, C., Tassy, O., Matifas, A., Molina, N., Charvin, G. eLife, 7:e34025, (2018) - DOI: 10.7554/eLife.34025
Controllable stress patterns in microfluidic devices Goulev, Y., Matifas, A. , Charvin, G. Methods Cell Biol., 147:29-40, (2018) - DOI: 10.1016/bs.mcb.2018.07.003
2017
Nonlinear feedback drives homeostatic plasticity in H2O2 stress response Goulev, Y., Morlot, S., Matifas, A., Huang, B., Molin, M., Toledano, M. B., Charvin, G. eLife, 6:e23971, (2017) - DOI: 10.7554/eLife.23971
2016
Kinetics of Formation and Asymmetrical Distribution of Hsp104-Bound Protein Aggregates in Yeast Paoletti C, Quintin S, Matifas A, Charvin G. Biophys J., 110(7):1605-14, (2016) - DOI: 10.1016/j.bpj.2016.02.034
2015
Two routes to senescence revealed by real-time analysis of telomerase-negative single lineages Xu Z, Fallet E, Paoletti C, Fehrmann S, Charvin G, Teixeira MT Nature Comm., 6:7680, (2015) - DOI: 10.1038/ncomms8680
Oscillatory Flow Modulates Mechanosensitive klf2a Expression through trpv4 and trpp2 during Heart Valve Development Heckel E, Boselli F, Roth S, Krudewig A, Belting HG, Charvin G, Vermot J. Curr Biol., 25(10):1354-61, (2015) - DOI: 10.1016/j.cub.2015.03.038
A quantitative approach to study endothelial cilia bending stiffness during blood flow mechanodetection in vivo Boselli F, Goetz JG, Charvin G, Vermot J. Methods Cell Biol., 127:161-7, (2015) - DOI: 10.1016/bs.mcb.2015.01.006
In silico control of biomolecular processes Uhlendorf J, Miermont A, Delaveau T, Charvin G, Fages F, Bottani S, Hersen P, Batt G. Methods Mol Biol., 1244:277-85, (2015) - DOI: 10.1007/978-1-4939-1878-2_13
2014
A memory system of negative polarity cues prevents replicative aging Meitinger F, Khmelinskii A, Morlot S, Kurtulmus B, Palani S, Andres-Pons A, Hub B, Knop M, Charvin G, Pereira G Cell, 159(5):1056-69, (2014) - DOI: 10.1016/j.cell.2014.10.014
Endothelial Cilia Mediate Low Flow Sensing during Zebrafish Vascular Development Goetz JG, Steed E, Ferreira RR, Roth S, Ramspacher C, Boselli F, Charvin G, Liebling M, Wyart C, Schwab Y, Vermot J. Cell Rep, 6(5):799-808, (2014) - DOI: 10.1016/j.celrep.2014.01.032
2013
Aging yeast cells undergo a sharp entry into senescence unrelated to the loss of mitochondrial membrane potential Fehrmann S, Paoletti C, Goulev Y, Ungureanu A, Aguilaniu H, Charvin G. Cell Rep, 5(6):1589-99, (2013) - DOI: 10.1016/j.celrep.2013.11.013
Pulse propagation by a capacitive mechanism drives embryonic blood flow Anton H, Harlepp S, Ramspacher C, Wu D, Monduc F, Bhat S, Liebling M, Paoletti C, Charvin G, Freund JB, Vermot J. Development, 140(21):4426-34, (2013) - DOI: 10.1242/dev.096768
Comparison of DNA decatenation by Escherichia coli topoisomerase IV and topoisomerase III: implications for non-equilibrium topology simplification. Seol Y, Hardin AH, Strub MP, Charvin G, Neuman KC. Nucleic Acids Res., 41(8):4640-9, (2013) - DOI: 10.1093/nar/gkt136
2012
Long-term model predictive control of gene expression at the population and single-cell levels Uhlendorf J, Miermont A, Delaveau T, Charvin G, Fages F, Bottani S, Batt G, Hersen P. PNAS, 109(35):14271-6, (2012) - DOI: 10.1073/pnas.1206810109
2011
Ultrasensitivity and positive feedback to promote sharp mitotic entry. Goulev Y, Charvin G. Mol Cell., 41(3):243-4, (2011) - DOI: 10.1016/j.molcel.2011.01.016