Unicellular organisms have evolved exquisite defense mechanisms to buffer against variations in internal physiological parameters or to counteract unpredictable environmental changes. The response of oxidative stress (e.g. hydrogen peroxide) is a fundamental and highly conserved regulatory system that allows the cell to ensure a precise redox balance, which is essential for proper physiological function. Yet, although the molecular players involved in the response are well characterized at the biochemical level, how these components integrate into a functional homeostatic system remains quite obscure.
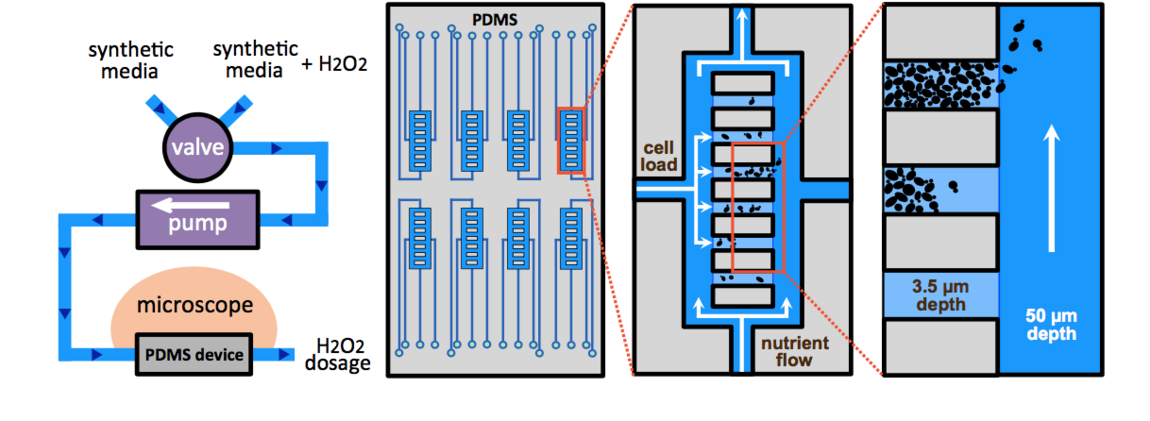
In this context, we have developed a microfluidic platform to monitor the physiological response of individual cells to various temporal stress patterns (i.e. steps, ramps, etc.) and we have deciphered the functional properties of the redox homeostatic system. We have shown that cell survival is highly stress-rate dependent: all cells consistently die when abruptly exposed to a given dose of hydrogen peroxide, yet can survive 10-fold higher doses when progressively exposed to the same stressor. This observation revealed that adaptation is limited by the response time of the homeostatic machinery, and unraveled an unprecedented ‘trainability’ of the cells to stress. We demonstrated that this particular feature, as well as the long described acquisition of tolerance, are mediated by key H2O2 scavenging enzymes called peroxiredoxins.
Following on this functional characterization of the response to oxidative stress, we are now trying to understand how cell reallocate their resources from cell growth to H2O2 scavenging when facing an oxidative stress challenge, knowing that some metabolites (e.g. NADPH) are involved in both anabolic processes and redox control.